Tag-LLM: Repurposing General-Purpose LLMs for Specialized Domains
ICML 2024
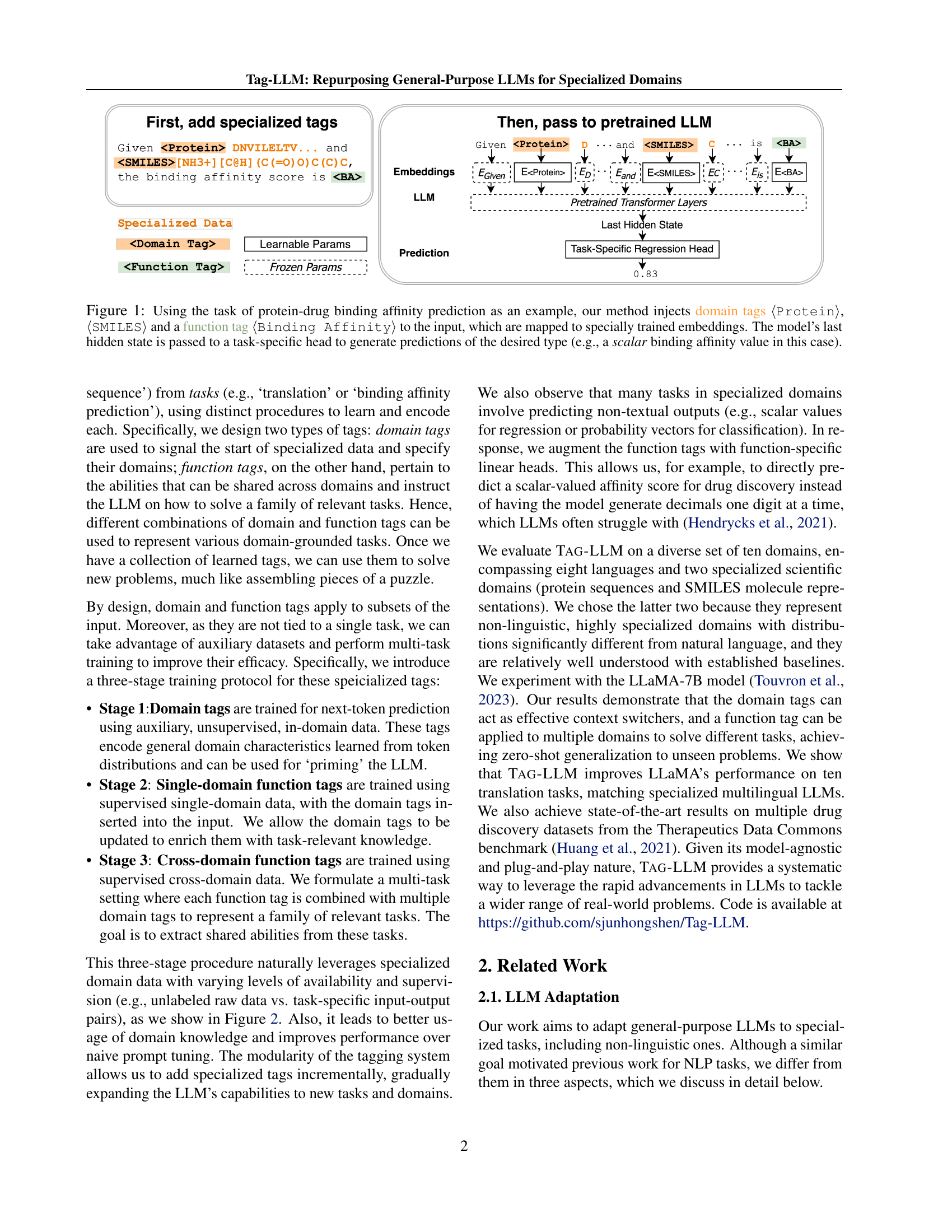
TL;DR
General LLMs struggle in specialized domains like chemistry and biology. We introduce learnable 'tags'—domain tags for context and function tags for tasks—that condition LLMs without retraining, enabling zero-shot generalization to new scientific problems.
Abstract
Large Language Models (LLMs) have demonstrated remarkable proficiency in understanding and generating natural language. However, their capabilities wane in highly specialized domains underrepresented in the pretraining corpus, such as physical and biomedical sciences. This work explores how to repurpose general LLMs into effective task solvers for specialized domains. We introduce a novel, model-agnostic framework for learning custom input tags, which are parameterized as continuous vectors appended to the LLM’s embedding layer, to condition the LLM. We design two types of input tags: domain tags are used to delimit specialized representations (e.g., chemical formulas) and provide domain-relevant context; function tags are used to represent specific functions (e.g., predicting molecular properties) and compress function-solving instructions.
Citation
@inproceedings{shen2024tagllm,
title={Tag-LLM: Repurposing General-Purpose LLMs for Specialized Domains},
author={Shen, Junhong and Tenenholtz, Neil and Hall, James Brian and Alvarez-Melis, David and Li, Liam},
booktitle={International Conference on Machine Learning},
year={2024}
}